
1Pass is a command line application that allows to explore 1Password OPVault format. Application was created because there is no official 1Password desktop client for Linux users. Only official desktop application allows for local passwords store in OPVault format. As a long term 1Password user, I don’t want to change my passwords manager for anything else only because I am working on Linux. For me it is really important to have choice where to store my passwords. I don’t feel comfortable with my passwords in the cloud. So here is the solution. Before I made the application, every time when I forgot password, I needed to use my phone to check how it goes in passwords manager. Now I can do it on a Linux PC. What is more, I can do it in Linux way – using CLI only.
Application is available only for Linux x86_64. Right now it is distributed as binary only. Installation process:
- Go to GitHub releases section and download the newest release.
- Extract downloaded archive in desired location.
- Run extracted binary.
If application do not run, it is probably permissions problem. Try chmod 755 1pass, this should resolve permissions problem.
For more comfy usage, binary can be added to $PATH (in .bashrc file):
EXPORT PATH=[path_to_binary_directory]:$PATH
Recommended way is to unpack downloaded archive in /usr/bin location. It will automatically make binary executable from terminal with typing just 1pass.
IMPORTANT: release 1.1.0 introduced update service for application (details below)
From release 1.1.0, application has implemented GitHub updates mechanism. Application automatically checks for new updates and notifies user about pending one. Application is not updating without user permission. To start update, run:
1pass update
It is recommended to give application root permissions during update because it is working on computer file system.
Whole update process:
- Check if there is new release on GitHub.
- Download newer release to temporary directory (with checksums).
- Extract downloaded archive.
- Compare checksums.
- Replace running binary.
- Clean cache (temporary files and directories).
From release 1.1.0, application has implemented interactive configuration tool. From release 1.2.0, application is prompting user for configuration on first run (the most important is default OPVault path to skip writing it ad-hoc). Whole configuration process relies on questions answering.
This is detailed description of all available settings:
1. Do you want to set default OPVault path? ([default_answer]) [y - for yes/n - for no]:
Default OPVault path ([previous_value]):
This setting allows to set default OPVault path. Configured path will be used by default if flag -v is not given to
command.
Default value: ""
2. Update notifications? ([previous_value]) [y - for yes/n - for no]: [value]
Decide if update notifications should be displayed. Type 'y' if yes or 'n' if no.
Default value: y
3. Update HTTP timeout in secods ([previous_value]) [1-30]: [value]
Set HTTP timeout for updates. This setting defines how long application should try to connect with GitHub for update
check. Slower internet connection will need bigger value. Value should be in range from 1 to 30 seconds.
Default value: 1
4. How often check for updates in days ([previous_value]) [0-365]: [value]
Set how often application should check for updates. Value is specified in days and should be in range form 0 to 365.
If 0 is set, application will check for update on every run.
Default value: 1
1Pass is a command line tool, so usage is limited to command variations. First of all type:
1pass
Command should launch application in GUI mode. Application can work in command line only mode also (without GUI). Provided commands:
1pass configure
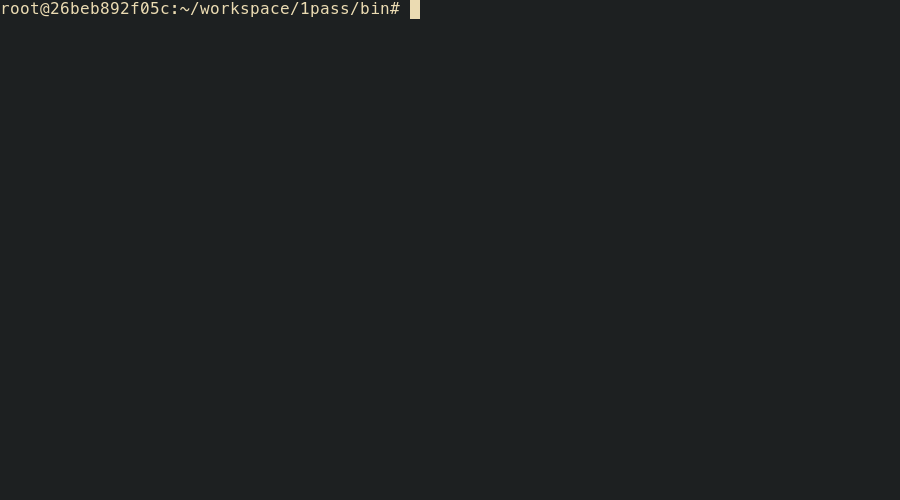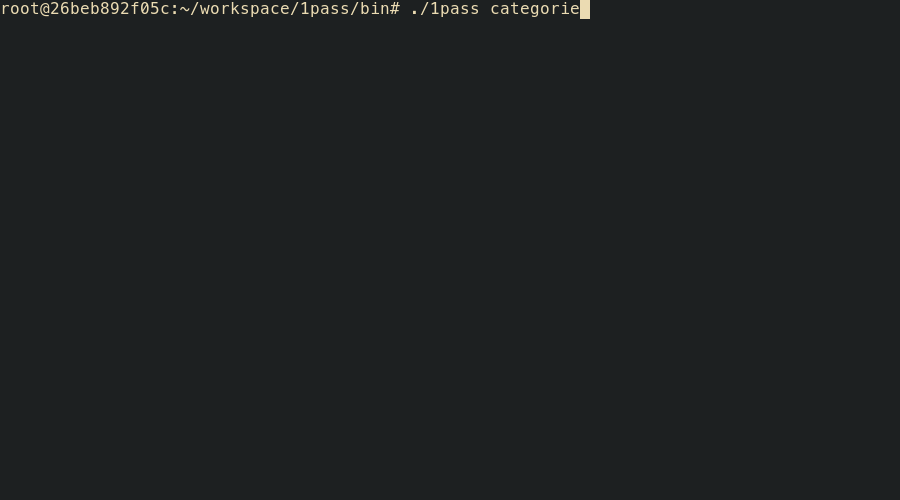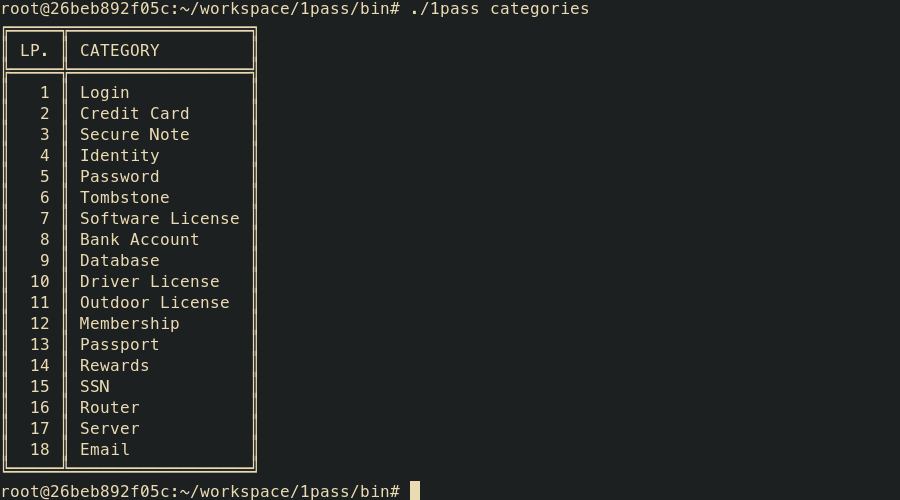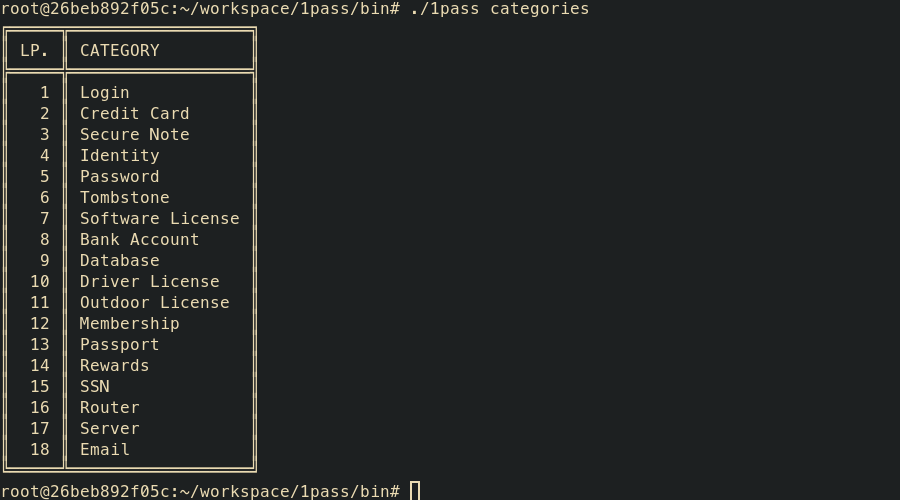
1pass categories
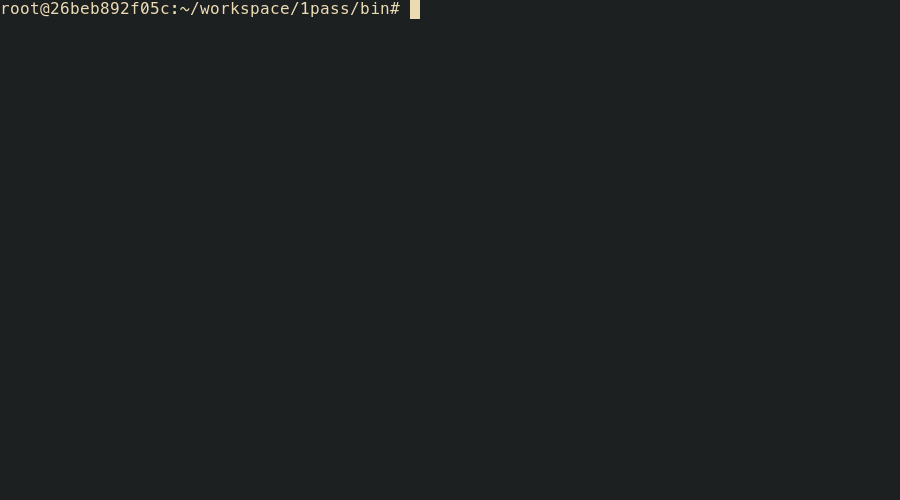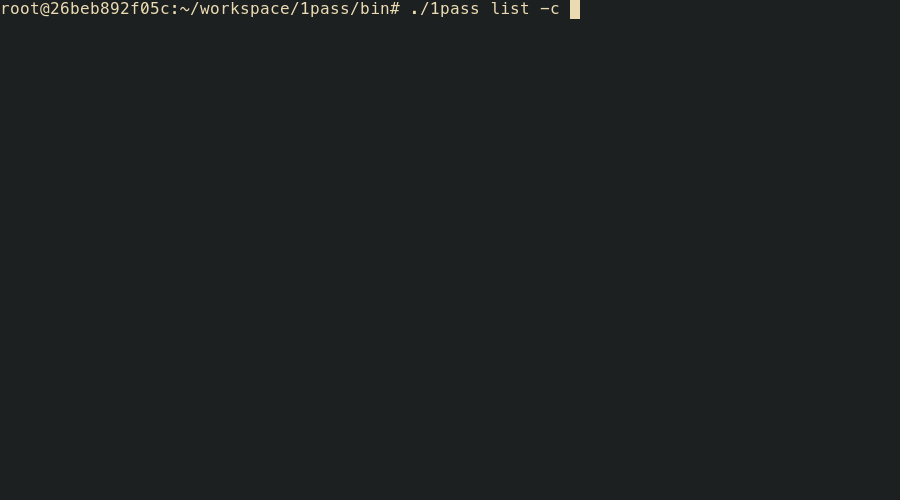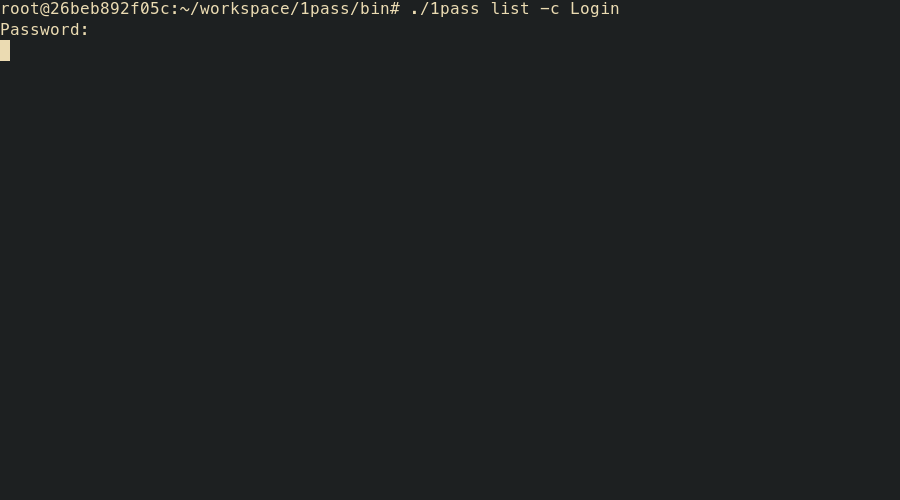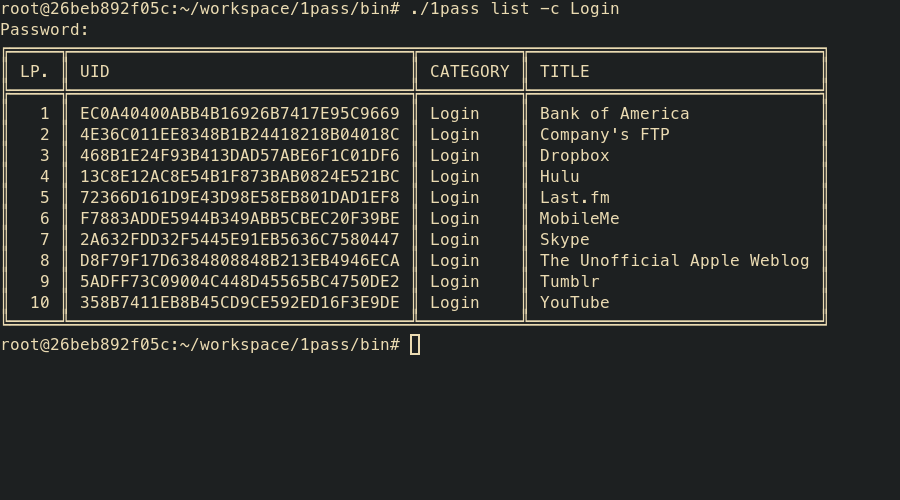
1pass list [-c <category>] [-n <name>] [-t] <path>
1pass overview [-t] <uid> <path>
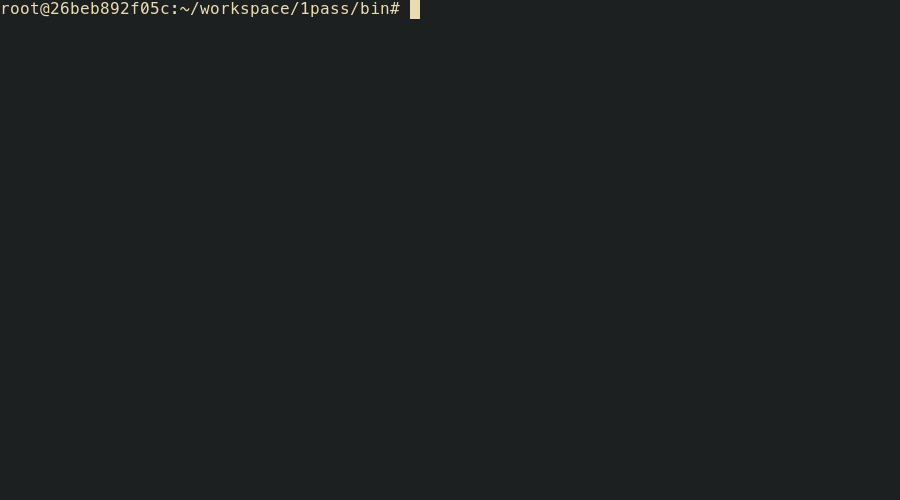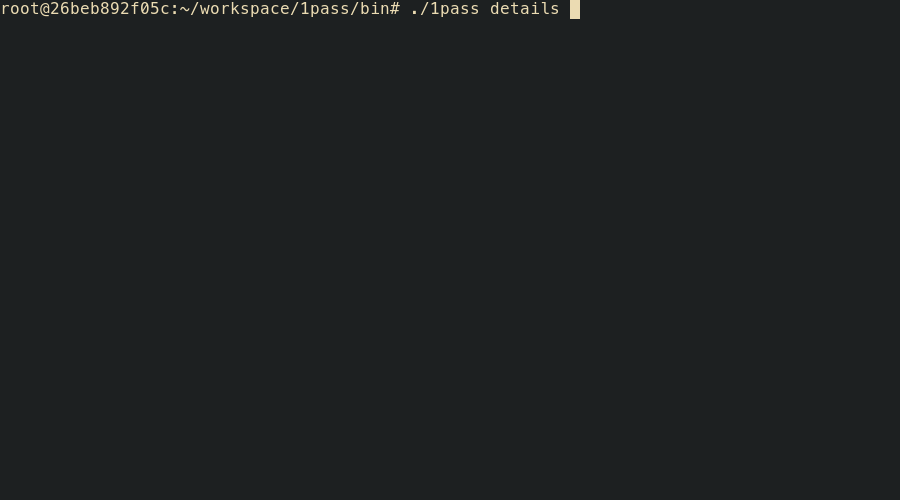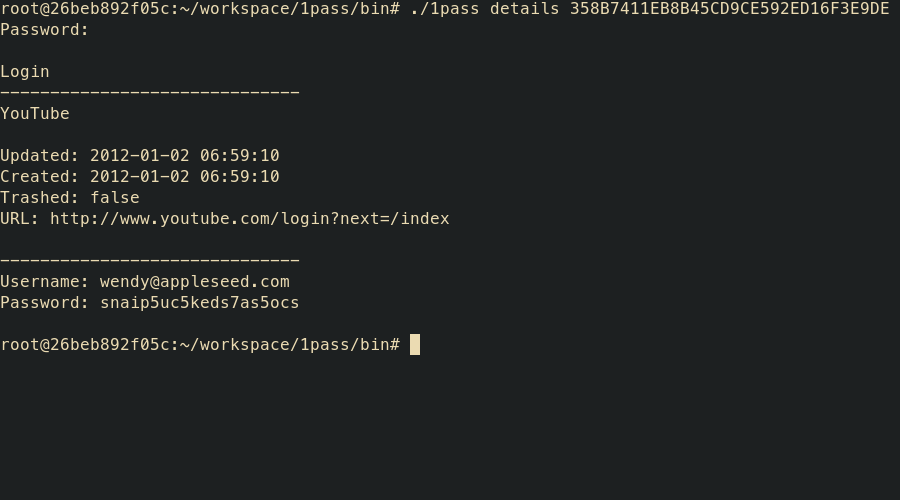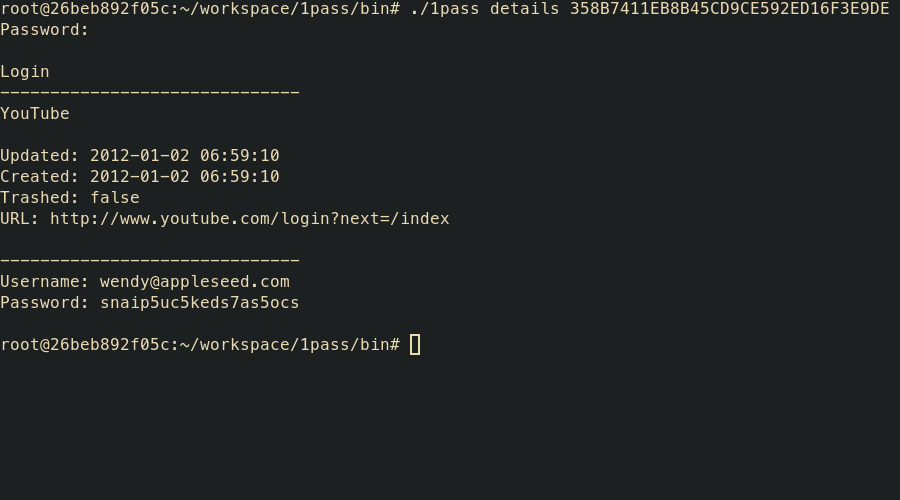
1pass details [-t] <uid> <path>
1pass update
1pass version
- configure – interactive application configuration (answer the questions), use help command to see what can be configured
- categories – display list of OPVault item categories (for filtering purposes)
- list – display list of items stored in OPVault
- overview – display overview of item without sensitive data
- details – display details of item with sensitive data
- update – check for update and upgrade 1pass
- version – check actual 1pass version
Legend:
- uid – unique UID of item (obtained with list command)
- path – path to 1Password OPVault
- -c – filter items over category
- -n – filter items over name/title
- -t – work on trashed items (archived)
- (FIX) [GUI] Notes padding for item details
- (FIX) [GUI] Notes padding for item overview
- (FIX) [GUI] Invalid password loop (application will not exit after displaying invalid password error)
- (FIX) [CLI] Inline update confirmation
- (FIX) [CLI] Notes padding for item details
- (FIX) [CLI] Notes padding for item overview
- (FIX) [CLI] Accept more reasonable update timeout during configuration (in range from 1 to 30 seconds)
- (FIX) [CLI] Update check can be shifted for one year maximum
- (FIX) [CLI] No configuration abort on invalid values (continue with actual state)
- (FIX) [API] Default update timeout set to 1 second
- (FIX) [API] Clear cache (temporary directory) before update
- (FIX) [API] Do not parse fields with value but without name
Versions of last five releases:
- 1.3.1
- 1.3.0
- 1.2.0
- 1.1.0
- 1.0.0
All actual work on project can be tracked in GitHub issues or GitHub projects.
Below you will find instructions how to contribute without changing my software developing workflow. All other type of contribution or contribution that do not follow rules, will be “banned” out of the box.
Who likes bugged software? Probably no one. If you will find any bug in application I will be really thankful. Bugs can be reported with GitHub issues. For the proper development cycle, I want to investigate reported bug. I also want to reproduce it and prepare technical description of issue to resolve it in next release. According to that, bugs should be reported with Bug issue template and bug label. If I will reproduce the bug and find fix for it, issue will be linked to issue with bugfix label – ready to work in next releases.
I am always opened for new ideas. New feature or change request can be reported with GitHub issues. There is special template named Request and request label. I will discuss this type of issues. If feature request/change is accepted, it will be linked with issue that has feature label – ready to implement in next releases.
It is really great if you want make some changes by yourself in my code base. First of all, some bureaucracy. Before you open issue, try to understand existing code. As you can see, this is multi module Go lang project in single GIT repository. Do you know why? I am really big fan of hexagonal software architecture because it is easier to control changes in external dependencies, core code base is a clear language (Go lang without dependencies), code is hermetic and easy to maintain. Even if it is overkill for small projects like this, it is my weapon of choice.
How architecture looks like right now?
- 1pass-core – core of the application (no external dependencies), business logic
- 1pass-parse – parsing component used to read data from OPVault format
- 1pass-up – application update component
- 1pass-term – component used to handle CLI interaction with application
- 1pass-app – real application (combines all of above)
Independent Go modules makes it easier to track changes than packages.
If everything is clear to this point and you still want to modify code, open an issue with template Bug (labels bug and pr) or Request (labels request and pr). The next step is to describe amount of work you want to do. The more detailed description, the better. Git branch name should follow the pattern:
<latest_release>/pr/<short_issue_title_with_underscores>/<issue_number>
1.0.0/pr/pretty_item_overview/#99
I am trying to use Conventional commits, so it is really important that your commits also should. Example:
feat(#11): get item overview
tests(#11): unit tests of get item overview
Look at the repository commits, you will handle it.
Every pull request will be discussed with me and merged to develop after acceptation (unit tests are welcome).
 https://github.com/MashMB/1pass
https://github.com/MashMB/1pass